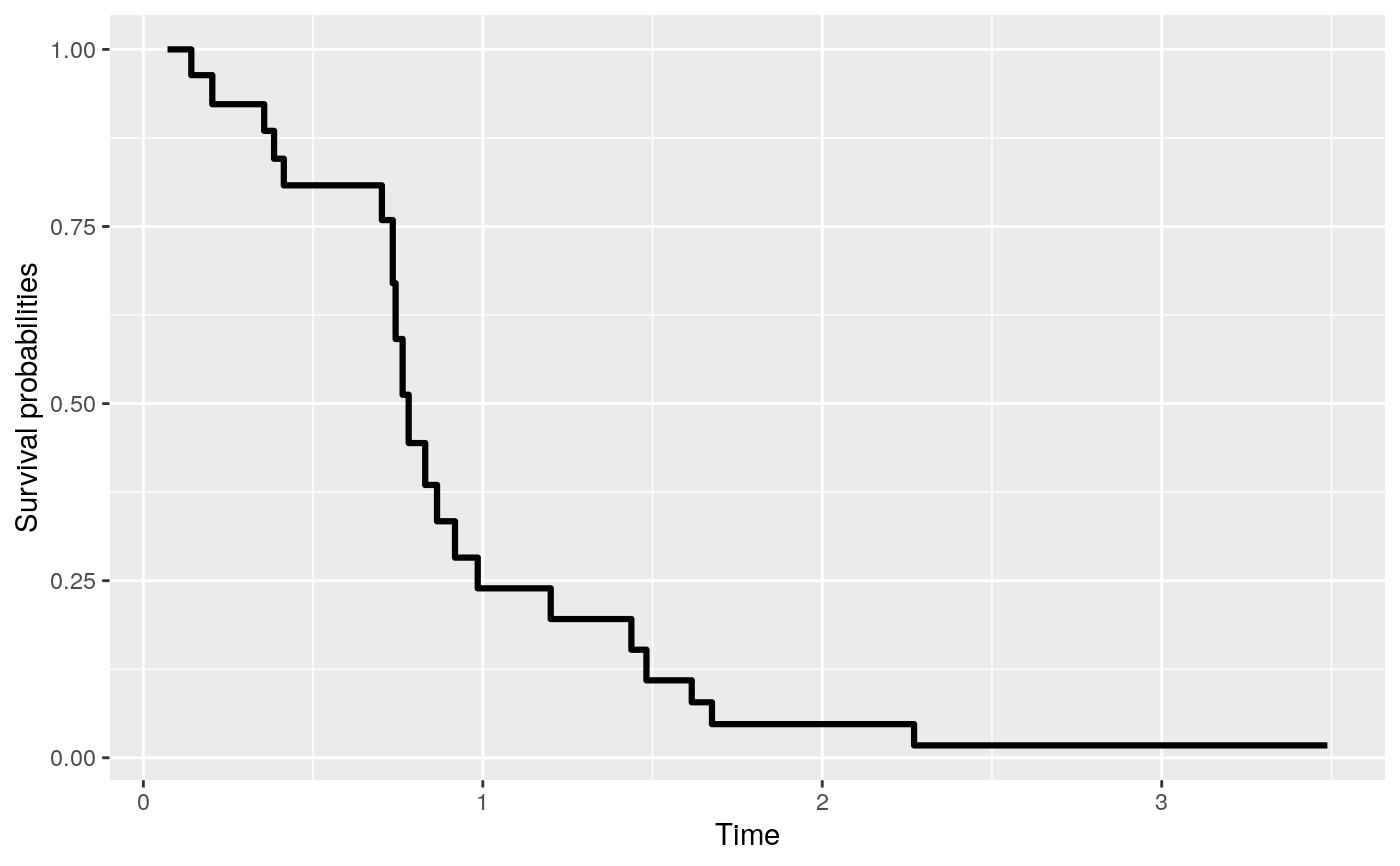
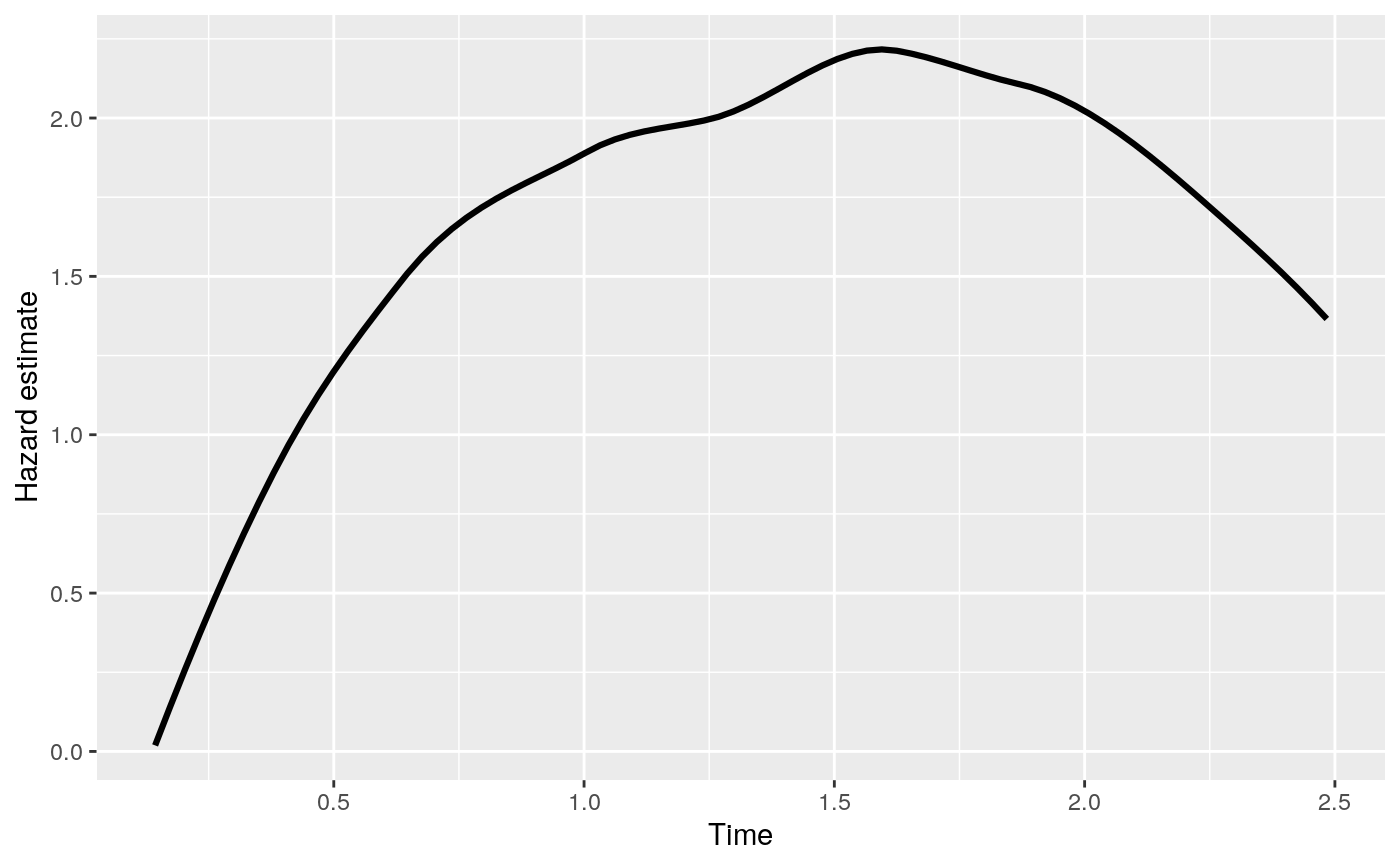
The function gives predicted values with a rocTree fit.
# S3 method for rocTree predict(object, newdata, type = c("survival", "hazard"), control = list(), ...)
Arguments
| object | is an |
|---|---|
| newdata | is an optional data frame in which to look for variables with which to predict. If omitted, the fitted predictors are used. If the covariate observation time is not supplied, covariates will be treated as at baseline. |
| type | is an optional character string specifying whether to predict the survival probability or the cumulative hazard rate. |
| control | a list of control parameters. See 'details' for important special
features of control parameters. See |
| ... | for future developments. |
Value
Returns a data.frame of the predicted survival probabilities or cumulative hazard.
Examples
data(simDat) fit <- rocTree(Surv(Time, death) ~ z1 + z2, id = id, data = simDat, ensemble = FALSE) ## testing data newdat <- data.frame(Time = sort(unique(simDat$Time)), z2 = runif(1)) newdat$z1 <- 1 * (newdat$Time < median(newdat$Time)) head(newdat)#> Time z2 z1 #> 1 0.07122299 0.80568 1 #> 2 0.08619285 0.80568 1 #> 3 0.11940614 0.80568 1 #> 4 0.12982798 0.80568 1 #> 5 0.14131532 0.80568 1 #> 6 0.14313760 0.80568 1