This function fits a linear regression model when there is a censored covaraite. The method involves thresholding the continuous covariate into a binary covariate. A collection of threshold regression methods are implemented to obtain the estimator of the regression coefficient as well as to test the significance of the effect of the censored covariate. When there is no censoring, the method reduces to the simple linear regression.
The model assumes the linear regression model: $$Y = a_0 + a_1X + a_2Z + e,$$ where X is the covariate of interest which is subject to right censoring, Z is a covariate matrix that are fully observed, Y is the response variable, and e is an independent randon error term with mean 0 and finite variance.
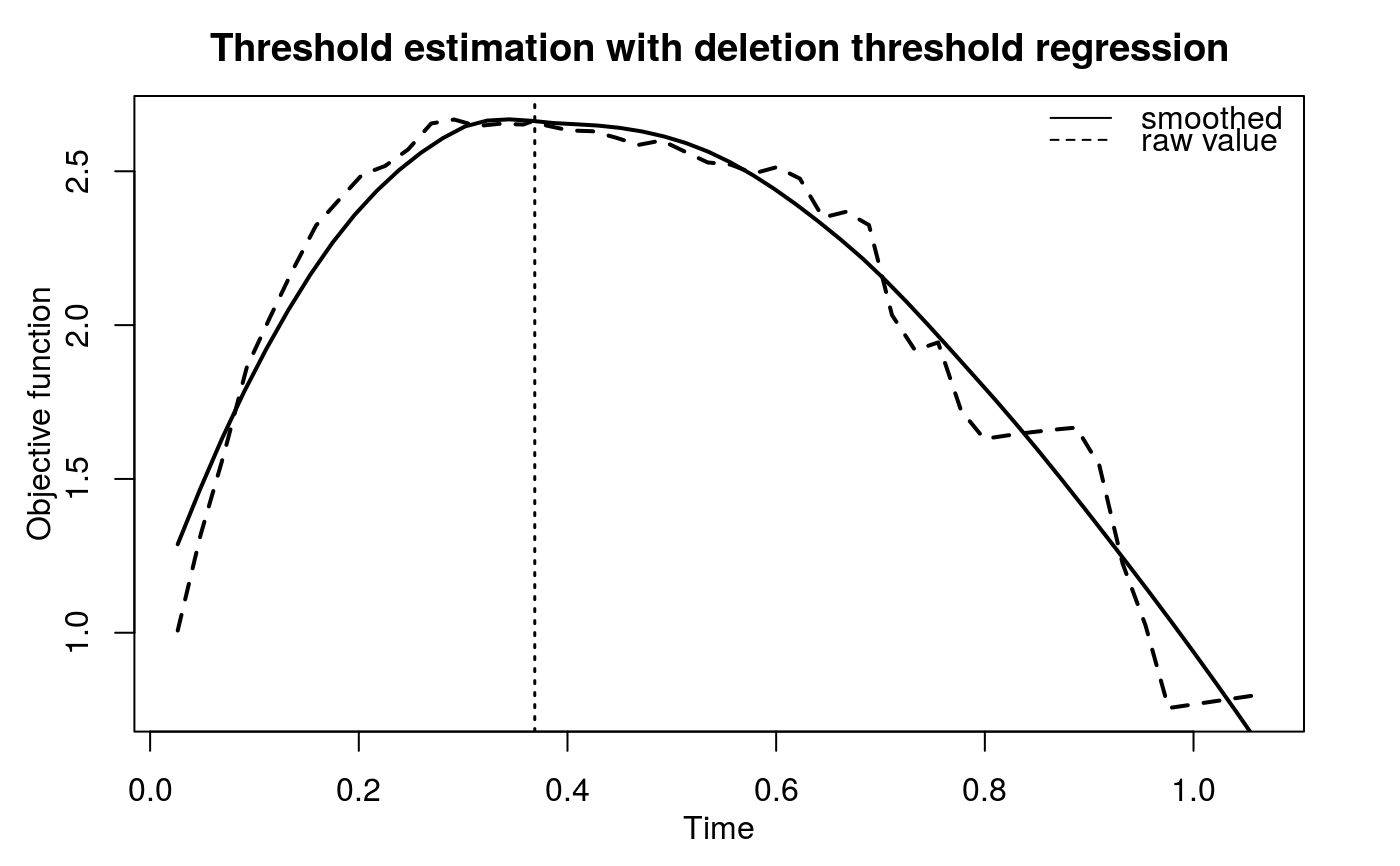
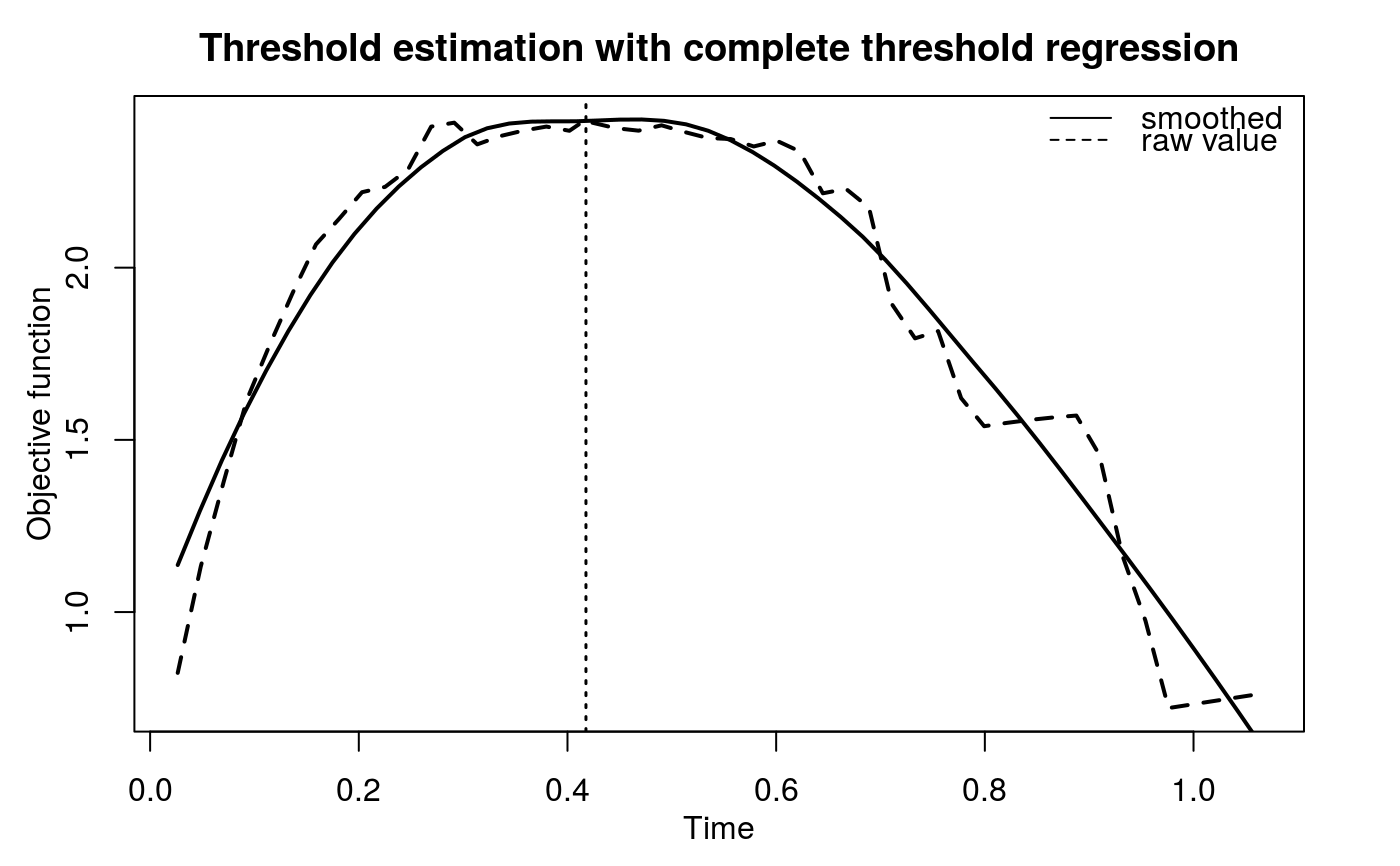
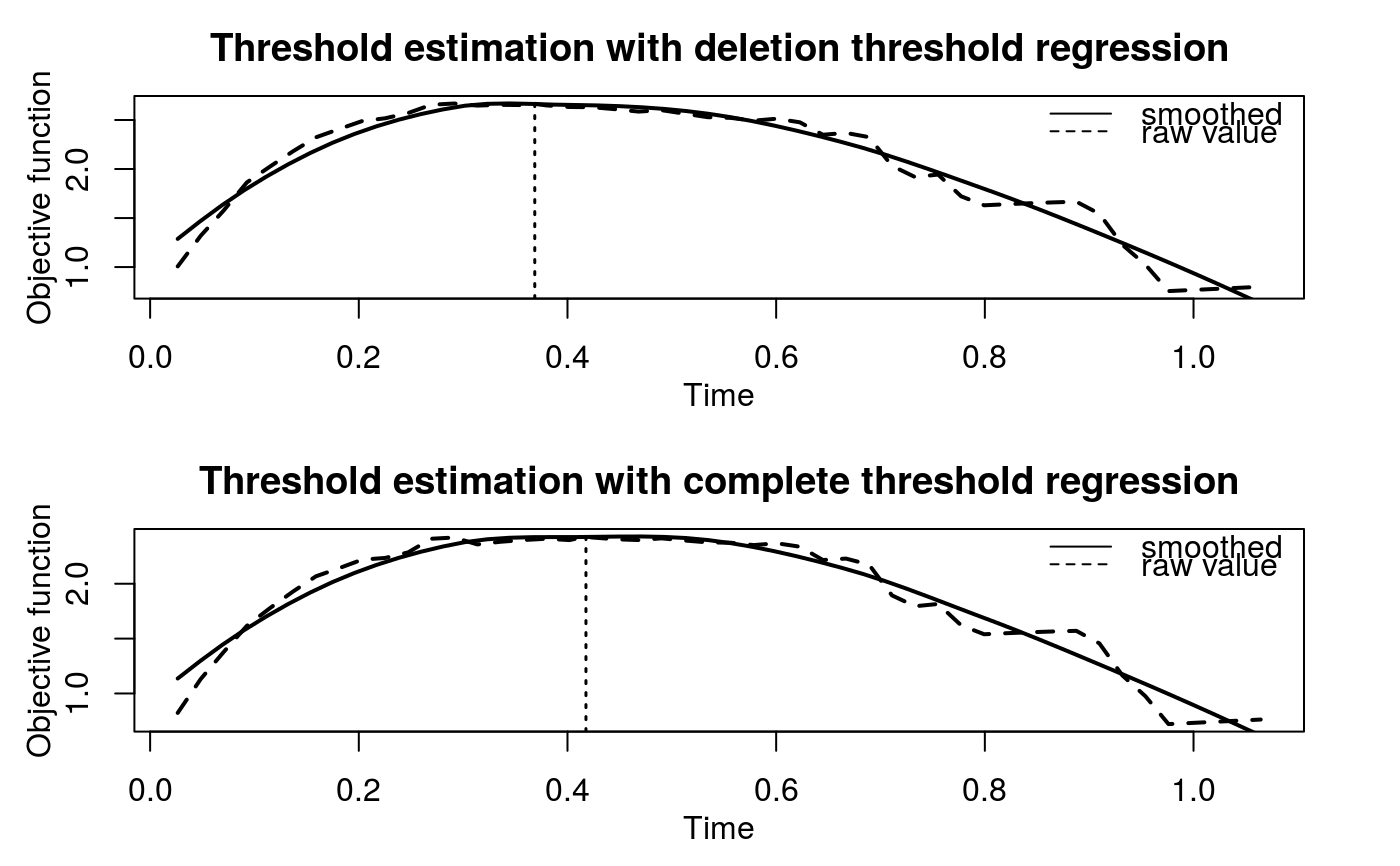
The hypothesis test of association is based on the significance of the regression coefficient, a1. However, when deletion threshold regression or complete threshold regression is executed, an equivalent but easy-to-evaluate test is performed. Namely, given a threshold t*, we define a derived binary covariate, X*, such that X* = 1 when X > t* and X* = 0 when X is uncensored and X < t*. The proposed linear regression can be expressed as $$E(Y|X^\ast, Z) = b_0 + b_1X^\ast + b_2Z.$$ The proposed hypothesis test of association can be tested by the significance of b1. Under the assumption that X is independent of Z given X*, b2 is equivalent to a2.
thlm(formula, data, method = c("cc", "reverse", "deletion-threshold", "complete-threshold", "all"), B = 0, subset, x.upplim = NULL, t0 = NULL, control = thlm.control())
Arguments
| formula | A formula expression in the form |
|---|---|
| data | An optional data frame list or environment contains variables in the |
| method | A character string specifying the threshold regression methods to be used. The following are permitted:
|
| B | A numeric value specifies the bootstrap size for estimating
the standard deviation of regression coefficient for the censored
covariate when |
| subset | An optional vector specifying a subset of observations to be used in the fitting process. |
| x.upplim | An optional numeric value specifies the upper support of the censored covariate. When left unspecified, the maximum of the censored covariate will be used. |
| t0 | An optional numeric value specifies the threshold when
|
| control | A list of parameters. The parameters are
|
References
Qian, J., Chiou, S.H., Maye, J.E., Atem, F., Johnson, K.A. and Betensky, R.A. (2018) Threshold regression to accommodate a censored covariate, Biometrics, 74(4): 1261--1270.
Atem, F., Qian, J., Maye J.E., Johnson, K.A. and Betensky, R.A. (2017), Linear regression with a randomly censored covariate: Application to an Alzheimer's study. Journal of the Royal Statistical Society: Series C, 66(2):313--328.
Examples
simDat <- function(n) { X <- rexp(n, 3) Z <- runif(n, 1, 6) Y <- 0.5 + 0.5 * X - 0.5 * Z + rnorm(n, 0, .75) cstime <- rexp(n, .75) delta <- (X <= cstime) * 1 X <- pmin(X, cstime) data.frame(Y = Y, X = X, Z = Z, delta = delta) } set.seed(0) dat <- simDat(200) library(survival) ## Falsely assumes all covariates are free of censoring thlm(Y ~ X + Z, data = dat)#> #> Call: thlm(formula = Y ~ X + Z, data = dat) #> #> Hypothesis test of association #> H0: a1 = 0, p-value = 0.0023 #>#> #> Call: thlm(formula = Y ~ Surv(X, delta) + Z, data = dat, method = "cc") #> #> Hypothesis test of association #> H0: a1 = 0, p-value = 0.0033 #>#> #> Call: thlm(formula = Y ~ Surv(X, delta) + Z, data = dat, method = "rev") #> #> Hypothesis test of association #> H0: a1 = 0, p-value = 0.0026 #>#> #> Call: thlm(formula = Y ~ Surv(X, delta) + Z, data = dat, method = "del") #> #> Hypothesis test of association #> H0: b1 = 0, p-value = 0.0080 #>thlm(Y ~ Surv(X, delta) + Z, data = dat, method = "com", control = list(t0.interval = c(0.2, 0.6), t0.plot = FALSE))#> #> Call: thlm(formula = Y ~ Surv(X, delta) + Z, data = dat, method = "com", #> control = list(t0.interval = c(0.2, 0.6), t0.plot = FALSE)) #> #> Hypothesis test of association #> H0: b1 = 0, p-value = 0.0040 #>## threshold regression with bootstrap thlm(Y ~ Surv(X, delta) + Z, data = dat, method = "del", B = 100)#> #> Call: thlm(formula = Y ~ Surv(X, delta) + Z, data = dat, method = "del", #> B = 100) #> #> Hypothesis test of association #> H0: b1 = 0, p-value = 0.0080 #> H0: a1 = 0, p-value = 0.0082 #>#> #> Call: thlm(formula = Y ~ Surv(X, delta) + Z, data = dat, method = "com", #> B = 100) #> #> Hypothesis test of association #> H0: b1 = 0, p-value = 0.0053 #> H0: a1 = 0, p-value = 0.0111 #>#> #> Call: thlm(formula = Y ~ Surv(X, delta) + Z, data = dat, method = "all", #> B = 100) #> #> Hypothesis test of association #> #> Complete-cases #> H0: a1 = 0, p-value = 0.0033 #> #> Reverse survival #> H0: a1 = 0, p-value = 0.0026 #> #> Deletion threshold #> H0: b1 = 0: p-value = 0.0080 #> H0: a1 = 0: p-value = 0.0090 #> #> Complete threshold #> H0: b1 = 0: p-value = 0.0053 #> H0: a1 = 0: p-value = 0.0136